Theoretical explanation of RdKit function-`AllChem.EnumerateStereoisomers(input)` for the generation of Stereoisomers.
Table of contents
Let's explore how the EnumerateStereoisomers function works at its core. It follows clear principles and a step-by-step mechanism, strictly adhering to specific rules to create valid isomers. I'll break down the process, explain the guiding rules, and highlight the rigorous validation steps involved
AllChem.EnumerateStereoisomers(input_molecule):
Basic's
Input: The function takes a SMILES string as input. SMILES stands for Simplified Molecular Input Line Entry System, and it's a compact text-based representation of a molecule's structure. You can find more information about SMILES here.
Parsing: The first step involves parsing the SMILES string into an RDKit molecule object. RDKit is a powerful toolkit for cheminformatics, and it allows for various operations on molecular structures.
Parameters Used:
include_cistrans: This parameter is used to specify whether to include geometric isomers. Geometric isomers have the same molecular formula but differ in the spatial arrangement of their atoms.include_optical: This parameter is used to control the inclusion of enantiomers. Enantiomers are mirror-image isomers of a molecule, often referred to as "left-handed" and "right-handed" versions.
Enumerating Stereoisomers: The core function,
EnumerateStereoisomers, performs the task of generating stereoisomers based on the input molecule and the specified parameters. Stereoisomers are isomers that have the same connectivity of atoms but differ in the spatial arrangement of those atoms.Geometric Isomers: If
include_cistransis set to true, the function will consider geometric isomers, which are isomers resulting from the restricted rotation around a bond. These isomers may have distinct spatial arrangements around double bonds or rings.Enantiomers: If
include_opticalis enabled, the function will also generate enantiomers. Enantiomers are non-superimposable mirror-image isomers and are relevant in the context of chirality.
Internal Working for Stereoisomers Generation:
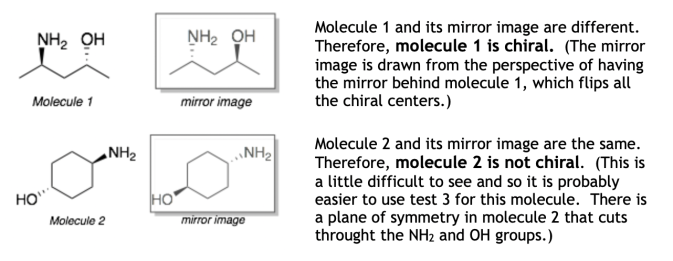
Chirality:
The function identifies molecules with chiral centers.Chiral centers are atoms with four distinct substituents.For each chiral center, the function attempts to create two configurations: R (right-handed) and S (left-handed).Configuration determination is based on theCahn-Ingold-Prelog priorityrules, ranking substituents by atomic numbers or mass.
Double Bonds:
The function considers molecules with double bonds, capable of cis-trans isomerism.It explores two possible configurations for double bonds: CIS and TRANS.This is relevant when there is restricted rotation around the double bond.
Combination of Configurations:
The function systematically combines different configurations of chiral centers and double bonds.Each isomer represents a unique spatial arrangement of atoms while maintaining the same molecular connectivity.By combining these configurations, the function generates a set of valid stereoisomers, each with distinct three-dimensional atomic arrangements.
Example:
ex: if you have 2 chiral centers & 1 double bond. there could be: 2 (R/S options for the first chiral center) 2 (R/S options for the second chiral center) 2 (cis/trans options for the double bond) = 8 possible combinations. 🤔
Next:
In the next Blog, I will go through the working of the code from the library rdkit, Chem based on their GitHub code.
This was the theoretical working of code.